Overview
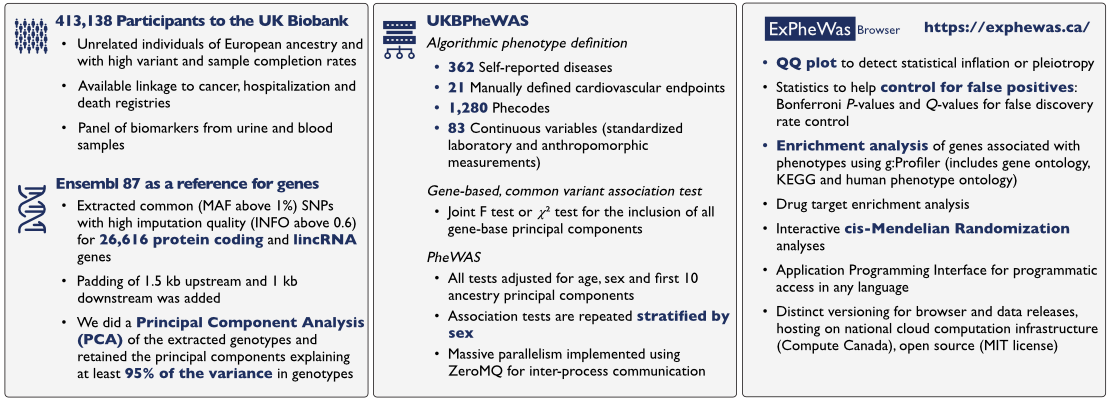
The ExPheWas browser showcases the results of a phenome-wide association study (PheWAS) using an approach that models the joint effect of variants at protein coding genes. In the current data release, we tested the association between 26,616 protein coding or lincRNA genes and 1,746 phenotypes available in the UK Biobank.
Briefly, we used a principal component analysis (PCA) of common variant genotypes to reduce the effective number of variables needed to capture statistical variance in the genotypes. Association testing is then done by assessing the improvement in goodness of fit from including the gene based principal components to a regression model. The experimental setup is described in more details in the figure below:
PheWAS results can then be browsed by gene or by phenotype.
Phenotype page
See all genes associated with a given phenotype.
List of phenotypes(or random example)
cis-Mendelian randomization
Estimate the causal (under the instrumental variable assumptions) effect of an exposure on an outcome based only on a single gene.
cis-MRCitation
If you use ExPheWas in your research, please consider citing:
ExPheWas: a platform for cis-Mendelian randomization and gene-based association scans
Legault, M.-A., Lemieux Perreault, L.-P., Tardif, J.-C., Dubé, M.-P.
Nucleic Acids Research, 2022. https://doi.org/10.1093/nar/gkac289
Browser release information
Version 1.3.0
Fixed (1 change)
A bug was fixed where the 95% confidence intervals for a cis-MR analysis via the API were incorrect. This bug did not affect the 95% confidence intervals displayed via the browser.
Data release information
We use distinct version numbers for the data release and the browser. This enables us to improve on the browser while unambiguously referring to the underlying data. You may refer to the data release version when reporting ExPheWas results and we are commited to maintaining stable access to previous data releases. There are currently two data releases (v0 and v1).
This browser uses data release v1.0 (2021-10-13)
This data release was presented at the International Genetic Epidemiology Society and the American Society for Human Genetics 2021 meetings.
Association testing was done using UKBPheWAS v2.0
Changes from the previous data release (v0)
- Sex-stratified analyses
- Interactive cis-MR
- Added lincRNA genes (Ensembl 87)
- Automatic enrichment analysis using g:Profiler (phenotype page)
- Asymetric padding of 1.5kb on the 5' end and 1.0kb on the 3' end when extracting genotypes for the PCA
- Phecodes were used instead of the raw ICD10 codes
- Genes on chromosome X
Support
For technical assistance or feature requests, we recommend you open a ticket on the project's Github page. For other inquiries, you may contact the corresponding author of the manuscript.